
You’ll need to adjust the path to reflect the location where the Excel file is stored on your computer (don’t forget to use double backslash within the path name to avoid any errors). Read_excel("C:\\Users\\Ron\\Desktop\\Products.xlsx") So this is the code to import the Excel file: library("readxl") The data that is stored in the Excel file is as follows: productįor demonstration purposes, assume that the file is stored under the following path: Let’s say that you want to import an Excel file into R (where the Excel file name is ‘Products’).
To start using the package installed, you’ll need to load it in the R Editor.įor instance, to load the readxl package, you’ll have to use: library("readxl") Step 4: Start using the package installed You may choose a mirror which is closer to your geographic location. Step 3: Select a Mirror for the installationįor the final step, select a Mirror for the installation.
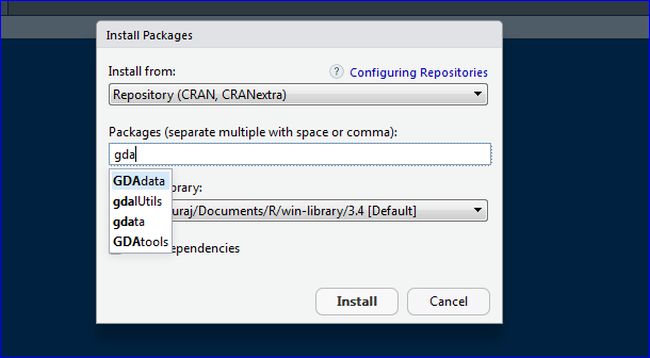
Once you are done typing the command, press ENTER to proceed with the installation. Now you’ll need to use the following template to install your package: install.packages("package_name")įor example, you may type the following command in the R Console in order to install the readxl package: install.packages("readxl") Lastly, the computers that I am working with on the cluster are Unix x86_64.> Step 2: Type the command to install the package In addition, I would also like to make sure that the packages are installed to a location of my choice since I do not have the permission to "write" in the default R directory (I believe that I can do this within R by using the. Ideally, I would like to download the packages files from CRAN to my computer, then upload these files to the cluster and install them using the appropriate commands in R. So in order to install packages I need to go to and manually download each one and their dependencies and install them in RStudio. Unfortunately the people in charge of my cluster are not being helpful in setting this up so I'm forced to consider this alternative approach. Note: I am aware that there is a way to avoid this issue by using an HTTP proxy as described in the R FAQ. R 3.4.0 will have tools::CRANpackagedb () to download the top-level PACKAGES.rds file (and of course you could just combine download.file () and readRDS () for that too). Since I am only using a few packages in my R code, I was hoping to avoid using the install.packages function by downloading and installing the packages manually. I am currently trying to run some R code on a computing cluster but cannot run the install.packages function due to some weird firewall settings on my cluster.


 0 kommentar(er)
0 kommentar(er)
